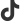Error: No such file or directory: '/outputpath/samplename/step3/filtered_feature_bc_matrix/barcodes.tsv.gz'
Release date : Jan 27,2026
- First step: Check the output directory. If raw_feature_bc_matrix exists and the files under this directory are not abnormally small, run the cell identification script for test:
Rscript /path/seeksoultools/lib/python3.XX/site-packages/seeksoultools/utils/cell_identify.R -i outputpath/step3/raw_feature_bc_matrix -e 3000
- If this script fails (e.g., due to a missing R package), then it’s probably environment issue. So run the source activate seeksoultools, or export PATH=/path/seeksoultools/bin:$PATH in order to activate environment, followed by rerunning the script.
- Tip: When installing seeksoultools, avoid creating soft links to another disk. Install it directly, ensure the environment name is unique, and use absolute paths when running scripts.
- If the files in the raw_feature_bc_matrix directory are abnormally small (e.g., only a few bytes), the GTF file may be problematic. For GTF format requirements, refer to the "Q&A 23: How to Build a Reference Genome?" section.
attachment:
Hot FAQ
Best practice for sequencing pooling and QC
Jan 27,2026
Jan 27,2026
Zero Expression of a Specific Gene in the Matrix
Jan 27,2026
Jan 27,2026
Enter your email address
Submit
Subscribe to receive updates on events, webinars, and product news.
For Research Use Only. Not for diagnostic use.
© 2026 SeekGene BioSciences. All rights reserved. | Privacy Policy
Top